What is AutoDock?

AutoDock is a suite of automated docking
tools. It is designed to predict how small molecules, such as substrates
or drug candidates, bind to a receptor of known 3D structure.
It has applications in:

AutoDock actually consists of three separate programs: AutoDock performs the docking of the ligand to a set of grids describing the target protein; AutoGrid pre-calculates these grids; and AutoTors sets up which bonds will treated as rotatable in the ligand.
In addition to docking, the atomic affinity grids can be visualised. This can help , for example, to guide organic synthetic chemists design better binders.
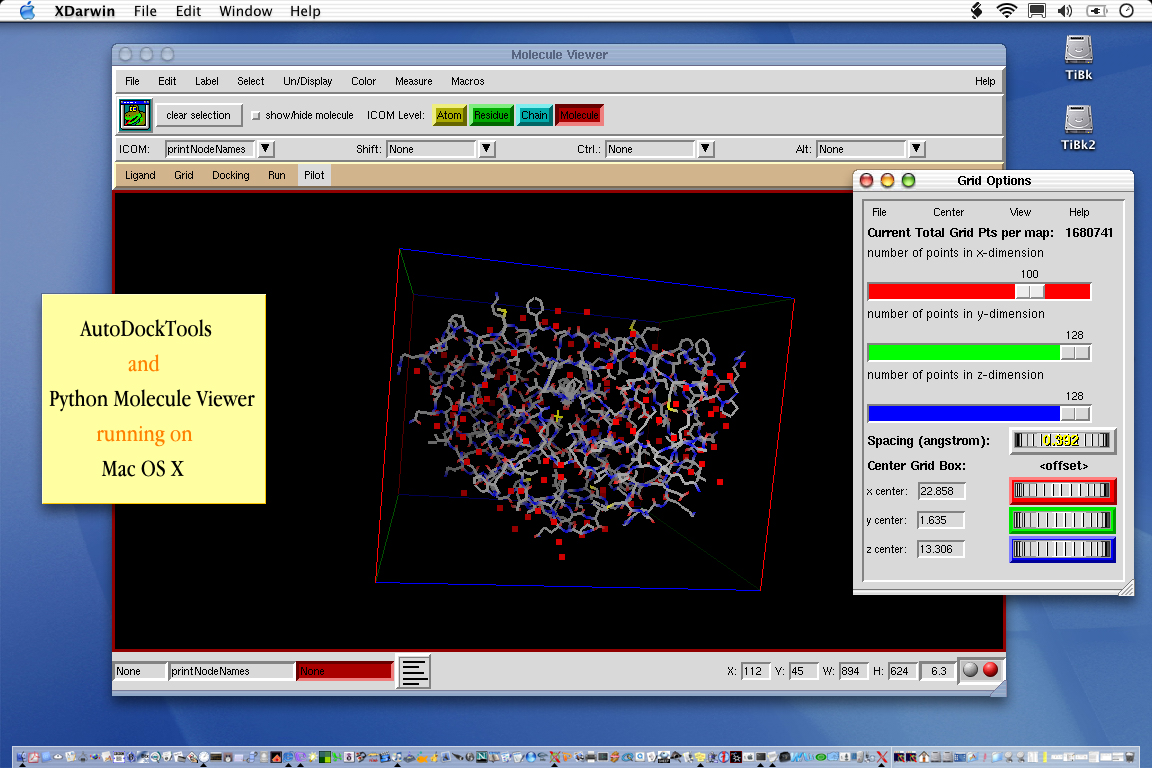
We have also developed a GUI called AutoDockTools, or ADT for short, which helps to set up and analyze dockings. We have an automated packager to help you download all the parts of ADT.
What's new?

AutoDock
3.0 is now out, and it is significantly faster than earlier versions.
This is thanks to improvements in the code, new algorithms and much more
efficient docking schedules than earlier ones. Rigid docking is blindingly
fast, and high-quality flexible docking can be done in around a minute.
Up to 40,000 rigid dockings can be done in a day on one cpu.
![]() AutoDock
3.0 now has a free-energy scoring function that is based on a linear
regression analysis, the AMBER force field, and a large set of diverse
protein-ligand complexes with known inhibiton constants. The best model
was cross-validated with a separate set of HIV-1 protease complexes, and
confirmed that the standard error is around 2 kcal/mol. This is
enough to discriminate between leads with milli-, micro- and nano-molar
inhibition constants.
AutoDock
3.0 now has a free-energy scoring function that is based on a linear
regression analysis, the AMBER force field, and a large set of diverse
protein-ligand complexes with known inhibiton constants. The best model
was cross-validated with a separate set of HIV-1 protease complexes, and
confirmed that the standard error is around 2 kcal/mol. This is
enough to discriminate between leads with milli-, micro- and nano-molar
inhibition constants.
AutoDock 3.0's search methods now include evolutionary methods, in addition to the Monte Carlo simulated annealing (SA) method of 2.4 and earlier. The Lamarckian Genetic Algorithm (LGA) is a big improvement on the Genetic Algorithm, and both genetic methods are much more efficient and robust than SA.
David S. Goodsell
Ruth Huey
William E. Hart
Scott Halliday
Rik Belew
Arthur J. Olson
News Flash! (6th
March 2002) We have just ported the graphical front-end
of AutoDock and AutoGrid, AutoDockTools, to Mac OS X!
Update: (10th
July 2002) Sorry, it is not yet available for downloading:
we are all at different conferences until the first week of August 2002.
When we all get back we will add ADT for Mac OS X to the distribution.
Thanks for all your support and enquiries about the OS X version. It is
going to be great!
Where is AutoDock Used?

AutoDock has now been distributed to nearly
1600 academic, governmental and non-profit institutions. About 1000
of these are the latest version, 3.0.5. A literature search of the ISI
Citation Index in May 2002 revealed more than 300 publications have cited
the AutoDock papers.
AutoDock 3.0 is also distributed to commercial enterprises: contact Art Olson at (858) 784-2526 for more information.
Why Use AutoDock?

AutoDock has been widely used and there are
many examples of its successful application in the literature (see References).
It is very fast, provides high quality predictions of ligand conformations,
and good correlations between predicted inhibition constants and experimental
ones.
For example, AutoDock Users Csaba Hetenyi and David van der Spoel just made the cover of July 2002's Protein Science.