In the late 1980's, as the first co-crystal structures of regulatory proteins bound to their target DNA sequences became available, it was hoped that some type of code linking protein structure to DNA specificity might be uncovered. For example, it would be gratifying to discover that arginine always chose to interact with guanine, while isoleucine specifically interacts with thymine. If that were the case, then it would be possible to rapidly develop design techniques for producing proteins that could bind any desired sequence of DNA. Of course, this has not happened. The nature of protein-DNA interactions is very complex, involving subtle changes in the conformation and hydration of both molecules which make predictive efforts at determining specificity quite difficult.

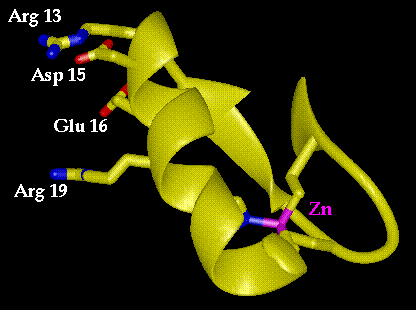
Figure 9.1 Structure of the first zinc finger of Zif268. Residues 13, 15, 16 and 19 are implicated in DNA recognition, in this case the base triplet GCG. The zinc ion is in the lower right portion of the structure and is chelated by two cysteines and two histidines.
One bright spot amid this complexity has arisen through structural studies of zinc fingers, a class of eukaryotic regulatory proteins. One particular class of these proteins, related to transcription factor IIIA from Xenopus (TFIIIA), has proven to be particularly intriguing. The crystal structure (solved by Carl Pabo and co-workers) of a complex formed between the DNA binding region of Zif268 (also known as Krox-24), a zinc finger protein found in mice, shows a modular pattern of DNA recognition.(1) The DNA binding portion of Zif268 is a small protein (only 90 residues) that contains three independent zinc finger domains of 24 residues each, connected by three to four amino acid linkers. Each finger consists of a single a helix joined to two strands of antiparallel sheet and held together via chelation of a zinc ion (Figure 9.1). The three zinc fingers bind to three adjacent three base sequences on the DNA target.
Interestingly, two of the three fingers have highly homologous sequences and bind identical three base sequences. The critical residues on the fingers that appear to determine this specificity are at positions 13, 15, 16 and 19 of a standard subunit sequence Pro1. (Tyr/Phe)2. X. Cys4. X. X. Cys7. X. X. X. Phe11. X. X13. X. X15. X16. Leu17. X. X19. His20. X. X. X. His24, where the conserved residues (such as Cys7 and Phe11) are involved in binding the Zn2+ ion or in preserving the hydrophobic core. Positions 13, 15, 16 and 19 are all on the face of the a helix that directly contacts the DNA. The sequence that binds GCG has the following substitutions: Arg13, Asp15, Glu16 and Arg19. To a first approximation, one might expect that any time you desire a protein that must bind, in part, a GCG sequence, that the above four residue combination will produce a zinc finger domain capable of filling that function.
Recognizing that these four positions are most critical in determining DNA specificity, Jeremy Berg and his co-workers at Johns Hopkins mounted a statistical search for other common groups of four residues at these four positions, in hopes of identifying other strong binding modules (2). In their search, they found that the following four residue combination was a common combination: Gln13, Ser15, Asp16 and Arg19. They created a mutant with this set of substitutions and discovered that, indeed, it does bind in a sequence specific manner, though not as might have been predicted. This exercise is intended to provide familiarity with the types of intermolecular contacts between protein and DNA and also with the limitations placed on "chemist's intuition."
Two files are needed for this exercise, entitled znfing.pdb and znDNA.pdb, (3) which are edited forms of the full PDB file 1zaa.full, containing the finger structure and the target DNA structure respectively. 1zaa.full may also be found in the directory. While you may be interested in inspecting the full structure, the edited versions allow the independent manipulation of the DNA and protein fragments with respect to each other.
Desjarlais and Berg prepared two mutant zinc fingers QSSE (Q13-S14-S15-E16) and QSSD (Q13-S14-S15-D16) from that are otherwise homologous to the zinc finger shown in Figure 9.1 (positions 13, 15 and 16 were modified). Of these two mutants, they report in their paper that the specificity of the mutant QSSD has a new specificity at the third position of the base triplet GCG. They conclude that the replacement of Arg13 by Gln13 is largely responsible for this change in specificity. Model the amino acid replacement and then make a base pair substitution for this guanine that looks chemically reasonable and identify a conformation of glutamine that can bind specifically to the new base pair.
Swapping glutamine for arginine can be done using the swapaa command (described in Exercise 3) and its conformation can be altered using the rot(ations) command. To substitute the base pair, you will need to use the "swapna' command.
will replace the base at position 3A with base "X", which should be a one letter code for one of the four DNA bases. The same command can be used to replace the second of the two bases in the pair as well. It should not be necessary to alter the conformation of the bases with respect to the phosphodeoxyribose backbone. However, you may wish to alter the position of the DNA duplex and the protein with respect to each other.
To move the two models independently of each other, select them individually in the Midas Control Panel, and then select them together when the proper relative positions have been obtained.
An interesting observation made in the course of this study was that it was necessary to make all three mutations at positions 13, 15 and 16 for the new zinc finger specificity to arise. The combination Q13, S15, D16 appears to be obligatory for activity. For example, mutant QSSE will not bind any DNA triplet. Apparently Glu16, though it doesn't make specific contacts to the DNA duplex, interferes in complex formation. Investigate the origins of this conflict by making the substitution at position 16 and considering the advantages of aspartate over glutamate at that position.
OK, so much for modeling... Experiment shows that the actual specificity of the QSSD mutant is GCT.(4) If this was not your initial guess, go back and investigate how thymine might productively interact with the glutamine at position 13.
(1) N. P. Pavletich and C. O. Pabo, "Zinc Finger-DNA Recognition: Crystal Structure of a Zif268-DNA Complex at 2.1", Science, 252, 809-817(1991).
(2) J. R. Desjarlais and J. M. Berg, "Redesigning the DNA-Binding Specificity of a Zinc Finger Protein: A Data Base-Guided Approach", Proteins, 12, 101-104(1992).
(3) The two DNA chains are labelled A and B in znDNA.pdb, and are numbered 1A-3A and 1B-3B. The protein chain has has no chain identifier. Its residues are numbered from 1 to 25.
(4)Proteins 13, 272 (1992)