Exercise Three looks at the dependence of secondary structure on sequence. In particular, certain residues are found unusually often at certain positions near the N-terminus and are believed to help stabilize the alpha helix. By modeling these substitutions, one can see how they provide structural stabilization to the helix.
This exercise provides more experience in manipulating dihedral angles, measuring distances and using vdw surfaces. In addition, this exercise introduces commands to swap amino acid side chains.
In the last exercise it was shown that only certain combinations of the dihedral angles phi and psi provide stable peptide conformations. An interesting extension of this observation is that two pairs of phi, psi values, when repeated along the length of the peptide chain, produce self-stabilizing structures referred to as secondary structure. The most commonly occurring conformations are known as alpha helices and beta sheet. Segments of the peptide chain that form secondary structures may be thought of as building blocks on the way to the construction of a larger proteins. In discussing the structure of globular proteins we will often think of it as agglomerations of a helix and beta sheet, connected by small stretches of "random coil", protein segments without the repetitive phi, psi values of secondary structures.
Despite the relative simplicity of secondary structures, until the mid 1980's, structural biochemists were frustrated by their inability to isolate stable alpha helices in solution (beta sheet is more complicated to isolate since each strand of sheet must pair with other strands, so that no individual strand can be isolated - by definition). It appeared that peptide sequences that form helices, while perfectly stable in a folded protein, could not be removed from that environment and be maintained in a helical state free in aqueous solution. Gradually, though, the understanding of helix stability increased as more crystallographic evidence was collected and solution studies became more sophisticated.
A particularly interesting analysis of crystallographic data was performed by Jane and David Richardson at Duke University.(1) Their study investigated the composition of 215 alpha helices from 45 different proteins in the PDB. They charted the positional frequency of each of the twenty amino acids in these helices and noted statistical deviations from random distribution. For example, it was found that glycine, a notoriously flexible amino acid, prefers to be found at either end of a helix, but not in the middle. Presumably, a glycine residue's inherent lack of conformational rigidity destabilizes the helix, which requires lock-step conformational regularity. In addition, the Richardsons also noticed distinct preferences for certain amino acids at the N-terminus of alpha helices. In particular asparagine and proline show marked preferences for specific positions at the N-terminus. Interestingly, glutamine is not found with any increased frequency at the N-terminal position, despite its structural similarities to asparagine. The focus of this exercise is to investigate the structural bases for these observations.
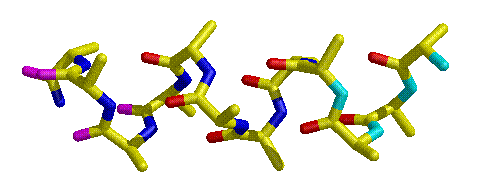
The alpha helix is held together in part by the glue of hydrogen bonds, with the amide C=O of the nth residue reaching ahead to hydrogen bond with the H-N of the n+4th residue (Figure 3.1). This works fine in the interior of the helix, but it means that the N-H groups at the N-terminus of the helix will be unpaired (as will the C=O groups at the C-terminus), since there are no helical residues four ahead of it. Therefore, the residues at the center of a helix are more likely to be in a stable conformation that those at either end. The Richardsons' work demonstrates that, in proteins, certain residues are prefered at the N-terminus of the helix because the structure of those particular amino acids stabilize the helical conformation.

Figure 3.1 Schematic of an alpha helix. The N-terminus of the helix is at the right. While the carbonyl oxygens and amide nitrogens in the center of the helix are colored the standard red and blue, respectively, the unpaired carbonyl oxygens are colored magenta at the C-terminus and the unpaired nitrogens are colored cyan, appearing at the N-terminus.
To understand their findings, a simple numbering scheme is introduced to identify position in the helix. The Richardsons defined the first residue in the sequence to have a (phi,psi) pair with the helix parameters as N1. The next residue is N2, followed by N3 etc. Ahead of N1 is the so-called N-cap residue. Its (phi,psi) pair is not necessarily that of a helix, but it is the first residue whose alpha carbon lies on the cylinder of the helix. Interestingly, 128 of the 214 alpha helices studied begin with one of the following four residues: glycine, serine, asparagine and aspartate. Glycine has already been discussed, but the striking information is that Asn is 3.5 times more likely to appear at the N-cap position than would be expected based on its overall representation in all helices. Asparagine is relatively uncommon in helices, so its predominant role at the N-cap position is worth considering, especially since the related amino acid residue, glutamine, only appears 40% as often as would be expected based on its overall inclusion in helices. Also, asparagine residues appear relatively rarely at the N1 position (70% of the random prediction). For some reason, it appears particularly desirable to place asparagine at the N-cap position.
Another interesting statistical anomaly occurs with proline, which appears only rarely in helices. However, in one position, N1, proline appears in 12% of all helices. Like asparagine, proline appears to have some unusually stabilizing influence at one particular position at the N-terminus of the helix. The goal of this exercise will be to explain why these two residues play such a significant role in stabilizine alpha helices.
Save the file ala.pdb to your working directory. These coordinates describe a single alanine residue, which will act as a seed residue for the construction of an alpha helix. After copying the file to your own directory, the file may be viewed in Midas by typing, in the console window:
It is possible to add amino acid residues onto the C-terminus of a peptide (or amino acid residue) using Midas. A 12 residue alpha helix will be the working model for this exercise and it needs to be built up on the single alanine in ala.pdb. This is possible using the "addaa" command:
This command indicates that Midas should add alanine as residue 2 in an alpha helix conformation (extended and beta sheet conformations are also available) to residue 1 in model #0. This command needs to be issued 10 additional times to build up a 12 residue helix. To avoid extreme tedium, simply edit the last command entered to increase the residue numbers by one each and hit return again. For example, the next entry would be:
To save the new model as a PDB formatted set of coordinates, type:
which saves the coordinates of model #0 to a file named helix.pdb in the working directory.
The amino acid residues in the new coordinate file are numbered in sequence from 1 to 12. On this numbering scheme, residue 1 is the N-cap residue, residue 2 is N1, residue 3 is N2, etc.The residues at the N-cap and N1 positions will be manipulated.
After Midas is open and the helix is colored, the first order of business is to replace residue 2 - or position N1, an alanine, with proline. To switch amino acids in the model, type the following command:
and this will "swap" proline for residue 2 of model 0. (Inserting the three letter abbreviation for any amino acid in for PRO will make the corresponding change.) Note that the new residue is uncolored. This can be remedied by applying "color byatom" again, or coloring it to a different set of specifications.
Note the steric environment around proline at position N1. Is it easily accommodated here? Check any clashes the proline side-chain may have with nearest neighbors either by monitoring distances or using vdw surfaces. (Note - pay close attention to CD of proline, which is unusual in being covalently attached to the amide nitrogen. How does proline at the position help alleviate the unpaired amide groups at the N-terminus?
After inspecting the N1 proline substitution, replace it again with alanine and then substitute proline for alanine at position N2. Why does proline occur only rarely at N2? Investigation of the steric environment around proline can be performed as described previously.
The Richardsons noted that asparagine chiefly hydrogen bonds to the NH group of residue N2 or residue N3, thus providing the stabilization that is otherwise lacking for the N-terminal amide nitrogens. Replace proline with alanine at position N2 and swap asparagine for alanine at the N-cap position.
Vary the conformation of Asn in the N-cap position, and verify that it can conveniently adopt conformations that will allow it to H-bond to the NH's of either N2 or N3. To monitor distances, the dist(ance) command is needed to follow distances between the appropriate atoms on the asparagine residue and in the helix backbone.
To manipulate conformation, bond rotations need to be assigned. For asparagine, three bonds require manipulation: ca-c, ca-cb and cb-cg. To activate them, the following three commands apply:
Note that "brot" designates back rotation, which holds the C-terminal part of the helix steady and rotates the part N-terminal to the designated bond. Appendix C contains some helpful guidelines for identifying appropriate hydrogen bonding geometry. When suitable geometries for H-bonding to the amide nitrogens of residues at positions N2 and N3 have been found, write down the appropriate rotation angles and the distances between hydrogen bond donor and acceptor. Also note any reservations regarding bad contacts elsewhere in the molecule. Finally, consider why it is that Asn is only able to take part in this kind of H-bonding from the N-cap position and not N1.
Delete the rotations from part 2 (using the ~rot command), swap glutamine for asparagine at the N-cap position, activate the four necessary rotations (note that glutamine has an extra methylene group and therefore one more bond to rotate). Determine if glutamine can H-bond to the amide nitrogens of the residues at positions N2 and N3 and note the results as for asparagine. Explain why it is that Gln appears relatively infrequently at the N-cap position. Take note of any bad conformations that might impede hydrogen bonding between Gln and the amide nitrogens of N2 and N3.
When finished, don't forget to quit Midas.
(1) J. S. Richardson and D. C. Richardson, "Amino Acid Preferences for Specific Locations at the Ends of a-Helices", Science (Washington, DC), 240, 1648-1652(1988).