|
|
|
|

|

|
Theoretical Biophysics Group, Dept. of Physics, UIUC
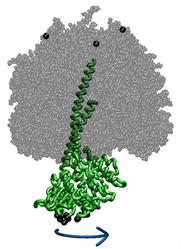 |
Every day, an active graduate student synthesizes her/his body weight
equivalent of the molecule ATP from a molecule ADP. The energy content
in ATP is used by the body to drive numerous biochemical syntheses as well
as mechanical processes like muscle action and brain processes. ATP
synthase contains two coupled molecular motors: one (F0) which
converts
an electrical membrane potential into a torque, the other (F1)
which converts a torque into chemical synthesis. Both motors are studied in
the Theoretical Biophysics Group through several approaches, quantum
electronics of ATP synthesis,
simulation of protein dynamics, and stochastic modeling of long-time
protein processes. The ATP synthase research team of the Theoretical
Biophysics Group seeks another graduate student to join this work.
|
|
The increase of computer power through parallel computing combined
with the strength of our group in molecular dynamics and graphics
programming has brought about a new approach to the study of the
molecular machines in living cells, interactive molecular dynamics.
This methodology permits researchers to visually connect to an ongoing
simulation of a biomolecular machine and manipulate it in real user
time in order to test hypotheses of its function. First research
projects are being accomplished now, one on studying the transport of
sugars through membrane channels, another to investigate the docking
of cytochrome c2 to the photosynthetic reaction center to donate an
electron. A new application will study the transport of chloride
through a membrane ion channel. The project provides a superb
opportunity in advancing computing technology, but also basic physics:
the manipulation generates irreversible work (IW) that needs to be
discounted to reveal the thermodynamic potential (TP) underlying the
process being studied. The possibility for the computation of TP from
IW has only recently been proven in an elegant theory on
non-equilibrium statistical mechanics that we will exploit.
|
|
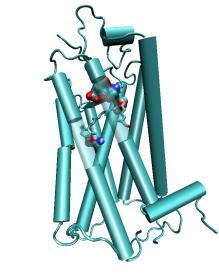 |
Since Newton, vision has attracted physicists seeking to explain how
light is sensed by organisms. Recently, the structure of a visual
receptor protein has been solved crystallographically (Science
289:739-745, 2000) and physicists are again called upon to
explain the fundamentals of vision, how quantum mechanics serves living
systems to see the world. The Theoretical Biophysics Group has studied
over the last twenty years a bacterial analogue of visual receptors,
bacteriorhodopsin, that actually uses light absorption to electrically
charge cell membranes. A new thesis project is based on a very recent
methodological breakthrough in our group: we are now capable to
calculate quantum-chemically the forces the visual chromophore retinal
experiences in the excited state reached after light absorption. This
can be done while the molecular dynamics simulation describes the motion
of the chromophore-protein system. We need to address now the
dynamics of jumps between excited and ground states at moments when
the two states approach each other energetically. Surface hopping poses
a fundamental problem in quantum mechanics closely related to the core
problem in quantum computing: the retinal at the moment of hopping is in
an entangled state (linear combination of ground and excited state) that
relaxes quickly to a non-entangled state. This relaxation process, due
to electron-phonon coupling needs to be properly described in the
framework of our new generation of photodynamics simulations.
|
Development of a Protein Scaffold System for Lipid Nanodisks
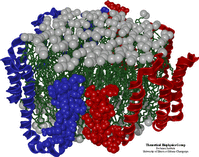 |
This is a close collaboration with the group of Steve Sligar, UIUC
Biochemistry. Based on previous work in the Theoretical Biophysics
Group and
recent advances in the Sligar group in designing a scaffold protein (SP)
from the Apo A1 motifs we will structurally model scaffold proteins
produced genetically in the Sligar group and possibly redesign them. We
will also investigate the immersion of membrane proteins in the lipid-SP
disks to guide and help analyze experiments. Lastly, we will attempt
to design lipid-SP systems that crystallize. This project offers an
opportunity for purely theoretical work, but also the fascinating
opportunity to combine experimental and theoretical work.
|
Single Molecule Electrical Recording
|
|
|