The substrate specificity of amino acyl tRNA synthetases (aaRS's) is absolutely critical in the process of translating nucleic acid sequence to protein sequence. While transcription can take advantage of standard base pairing and the ribosome does likewise, no such simple schemes are available to the enzymes that must join the correct amino acid to its corresponding tRNA molecule. An aaRS must be able to recognize, with absolute specificity, the structure of a given class of tRNA molecules out of twenty different isoacceptor groups (an isoacceptor group consists of one or more tRNA's that each accept the same amino acid) as well as the structure of the amino acid that is to be loaded. It would be certain death for an organism if, for example, seryl tRNA synthetase (SerRS) were to casually replace serine with alanine onto tRNA(Ser). Since the ribosome does not screen for correctly loaded tRNA molecules, this mistake would introduce alanine into the wrong positions of a growing peptide chain.

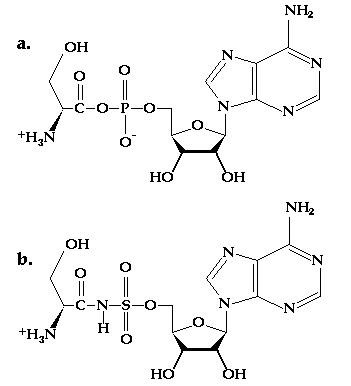
Figure 10.1 (a) The structure of seryl adenylate, a substrate for SerRS. (b) The structure of a sulfamoyl inhibitor of SerRS. The N-S bond is less susceptible to cleavage than the carboxylate-phosphate anhydride.
To investigate the means by which one particular aaRS performs these dazzling acts of substrate screening, Stephen Cusack and co-workers have performed crystallographic analyses on SerRS bound to analogs of seryl adenylate (Figure 10.1a) and to tRNA(Ser).(1,2) The complex of SerRS with tRNA(Ser) provided some novel results. Previous studies of GlnRS and AspRS bound to their respective tRNA's showed that the specificity resulted largely from direct interactions between base pairs on the tRNA molecule and amino acid side chains from the enzyme. In the case of SerRS, it has been known for some time that all members of the serine isoacceptor group possess an unusual D-loop, with a deletion at position 17 as well as extensive insertions (it is 20 bases longer than in other tRNA's). These changes lead to an unusual tertiary structure in tRNA(Ser) molecules. SerRS recognizes tRNA(Ser) chiefly through backbone interactions that are only available from this tertiary structure, and only makes secondary contacts to specific nucleotide bases.
The question of how SerRS recognizes serine specifically is the issue to be addressed by this exercise. The challenge that SerRS faces is to be able to distinguish serine apart from cysteine, threonine, alanine and glycine. Cysteine and threonine are challenges because they each possess similar functional groups to serine's. Alanine and glycine are problematic, on the other hand, because any pocket that could accept serine would also be able to accept those smaller groups. The crystal structure of a seryl adenylate analog (Figure 10.1b) bound to SerRS provides a starting point for this inquiry. By substituting the other four amino acids for serine in the structure of the inhibitor you will be able to identify the interactions SerRS makes to serine that exclude other amino acids from binding equally well.
The file 1set.pdb contains some modifications from the original pdb file (1set.full). SerRS appears as a dimer in the crystal structure, but in the modified file only one monomer is present, and the waters of hydration have been removed. In addition, there has been some light editing on the ATOM lines for the inhibitor to make amino acid substitutions possible.
To identify how SerRS specifically binds serine in its active site, study the serine binding pocket. By investigating the enzyme active site environment around the inhibitor (residue 422*-423*) it should be possible to identify the specific contacts made to the substrate's side chain. Prepare a list or table of those contacts and describe their importance in determining specificity. The next step is to use the swapaa command to replace serine with each of the following amino acids: alanine, cysteine, glycine and threonine. In each instance consider the contacts made by the side chains of these amino acids in the binding pocket. It may be necessary to adapt rotation angles about the Ca-Cb bonds to search for a "best" orientation.
(1) H. Belrhali, A. Yaremchuk, M. Tukalo, K. Larsen, C. Berthet-Colominas, R. Leberman, B. Beijer, B. Sproat, J. Als-Nielsen, G. Grubel, J.-F. Legrand, M. Lehmann and S. Cusack, "Crystal Structures at 2.5 Angstrom Resolution of Seryl-tRNA Synthetase Complexed with Two Analogs of Seryl Adenylate", Science, 263, 1432-1436(1994).
(2) V. Biou, A. Yaremchuk, M. Tukalo and S. Cusack, "The 2.9 Å Crystal Structure of T. thermophilus Seryl-tRNA Synthetase Complexed with tRNA(Ser)", Science, 263, 1404-1410(1994).