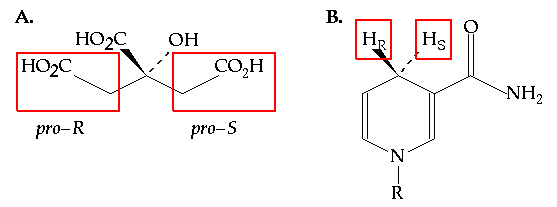
One of the most subtle stereochemical choices made by enzymes relates to their ability to distinguish between two identical atoms or functional groups bonded to a common carbon atom. For example, it was long thought that enzymes would be unable to distinguish between the two carboxymethyl groups of citric acid (Figure 7.1A). The ability to make this subtle distinction relies on stereochemistry. Two enantiotopic or diastereotopic groups are readily distinguished in the chiral environment of an enzyme active site. This understanding did not arise until 1948 (1) but it was rapidly appreciated by the biochemical community and other examples of stereoselectivity between stereotopic groups were rapidly identified. Perhaps none, however, has been more thoroughly studied since that time than the stereoselectivity shown by dehydrogenases for one of the two diastereotopic hydrogens at C4 (Figure 7.1B) of NADH's dihydronicotinamide ring.

Figure 7.1 (A) The structure of citric acid. The two carboxymethyl groups are enantiotopic and are indistinguishable in an achiral environment. (B) The structure of NADH (R = ADP ribose). The two hydrogens at C4 of the dihydronicotinamide ring are diastereotopic because there are other stereocenters in the molecule. Hr is the pro-R hydrogen and Hs is the pro-S hydrogen.
About one half of the dehydrogenases studied to date show pro-R specificity (that is they transfer the pro-R hydrogen) while the other half are pro-S specific (no known dehydrogenase is stereorandom). The basis for this stereospecificity is clearly based on the structure of the NAD(P) binding site. Depending on how the dihydronicotinamide ring is bound in the active site, either the pro-R or pro-S hydrogen will be positioned towards the substrate that is to be reduced. Note, however, that there is a great deal of structural conservation among NAD(P) dependent dehydrogenases. The dinucleotide binding domain is a common motif among of these enzymes and the NAD(P) cofactor tends to be bound in a common manner.(2) The one significant difference among these enzymes is the orientation of the nicotinamide ring itself. It can be bound in either the syn- or anti- conformations, which are related by a 180 degree rotation about the glycosidic bond linking the nicotinamide or dihydronicotinamide ring to the C1 of ribose (see Figure 7.2). Benner postulated (3) that all dehydrogenases that are pro-R specific will bind NADH with the dihydronicotinamide in the anti- conformation and pro-S enzymes will bind NADH in the syn- conformation, because these respective conformations will direct the correct hydrogen towards the substrate.
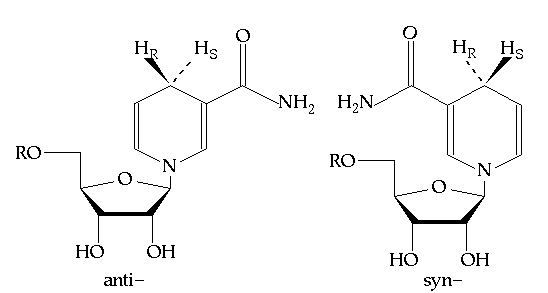
Figure 7.2 The anti- and syn- conformations of NADH (R = ADP).
The goal of this exercise is to test this postulate by inspecting a crystal structure of a binary complex of a dehydrogenase with cofactor, or a ternary complex with both cofactor and substrate bound in the active site. Inspection of the nicotinamide/ dihydronicotinamide ring (depending on whether it is the oxidized or reduced form of the cofactor present) will allow the determination of which side is oriented towards the substrate (or substrate binding pocket if no substrate is present).
In this exercise PDB files will be accessed directly from the Brookhaven National Laboratory WWW site, using Netscape to connect to the PDB home page.
The goal is to find a PDB entry that contains NAD(P) bound to the active site of a dehydrogenase, so that the conformation of the bound cofactor can be examined. The easiest way to find such an entry is to search the bibliographic entries for the keyword "NAD", which will bring up all structures containing either NAD or NADH bound to an enzyme. Simply click on the "Bibliographic header keyword search" link. Feel free to choose any entry, but be careful to read the bibliographic entry (with the suffix ".biblio") for the structure before deciding to download the coordinate file (with the suffix ".full") over the internet. Downloading such a large file can be somewhat time consuming, and you want to be sure that it is actually a file that will be of some use . When the file has been accessed, simply save it to the working directory in preparation for examining the structure in Midas.
Open the chosen file in Midas and color it as per usual. Since this will be a full protein model, it will be extremely complex and it will be useful to take steps to simplify it so that the image can be examined more closely for the desired information. The easiest way to locate the cofactor in this structure is to type
and only NAD(P)(H) will be displayed. There may be more than one molecule on the screen if the protein in question is a dimer or tetramer. In any case, you will be interested in the environment directly surrounding the cofactor, which can be examined by typing:
and then pick atom C4 of the (dihydro)nicotinamide ring. Only those residues within 7 A of C4 will be shown.
What is the conformation of NAD bound to the active site? Report the dihedral angle defined by the ring oxygen and C1 of ribose, the ring nitrogen and C2 of the nicotinamide ring. Identify which face of the ring is directed towards the substrate binding pocket and predict the stereospecificity of the reaction with respect to NAD(P). Does this agree with Benner's postulate?
What residues are responsible for maintaining the syn- or anti- conformation of the nicotinamide ring. Identify those residues that form stabilizing interactions with the C3 carboxamide group. If the nicotinamide ring were to rotate by 180 degrees about the glycosidic bond, would unfavorable interactions (steric clashes) result? Identify them.
(1) A. G. Ogston, "Interpretation of Experiments on Metabolic Processes, using Isotopic Tracer Experiments", Nature, 162, 963 (1948).
(2) See Branden and Tooze, Introduction to Protein Structure, Ch. 10, Garland Publishing, 1991
(3) K. P. Nambiar, D. M. Stauffer, P. A. Kolodziej and S. A. Benner, "A Mechanistic Basis for the Stereoselectivity of Enzymatic Transfer of Hydrogen Atom from Nicotinamide Cofactors", J. Am. Chem. Soc., 105, 5886 (1983)